Draw The Structure Of Ethanol Molecule
Building Molecules¶
In the first tutorial you have learned how to construct a molecule by building it out of atoms. That may be a complex task for bigger molecules. AMSinput has other ways to build molecules.
The quickest is to search ( ) for a molecule inside AMSinput, and use it if it is available.
) for a molecule inside AMSinput, and use it if it is available.
Another way is to search for the molecule on the Internet, and use either the xyz coordinates or the SMILES string
Or you can build it using the structure tool in AMSinput. As a more realistic example using the structure tool, you will build a small peptide chain. Then you will learn how to use the predefined metal complex structures. You will also learn how to set up your own structures library.
Finally, you can combine the crystal tools to cut molecular systems out of crystals. As an example we will make a sphere of Cu atoms.
Start AMSinput¶
For this tutorial we again prefer to work in the Tutorial directory:
-
Start AMSjobs
Click on the Tutorial folder icon
Start AMSinput via SCM → New Input
Search for ethanol¶
The quickest solution: search (and find) ethanol.
-
Press ctrl/cmd-F to activate the search box (or click the search icon
 in the panel bar)
in the panel bar)Enter 'ethanol' as search text (without quotes)
Click on the 'Ethanol (ADF)' match
Rotate to get a good view

Your ethanol is ready. The (ADF) in the search results mean that the molecule has already been optimized by ADF, using the BP86 XC potential with a TZP basis set and small core.
For the next demonstration we need a file with the xyz coordinates of ethanol. You can make such a file using the Export Coordinates option:
-
Use the File → Export Coordinates… menu command
Enter 'example.xyz' as filename and make you are saving to the Tutorial folder
Click Save
Import XYZ for ethanol¶
To import a molecule if you have its structure as xyz file (with element types), you can either use the File → Import Coordinates… or the Edit → Paste command.
-
Use the File → Close menu command in AMSinput
Click 'No' when asked if you want to save your changes
Click in the AMSjobs window to activate it
Enable Filter → Other (if it is not yet enabled)
Use the Job → Refresh List menu command (or press F5)
Click the triangle in front of example to show the example.xyz file
Double click on the .xyz file (listed in Local files)
AMSinput will start and automatically import the .xyz file.
Tip
Copy/Paste data into AMSinput: works for many formats (like xyz, SMILES or InChI strings)
An alternative and often easier way: just copy the contents of the .xyz file.
The coordinates are:
C 0.01247000 0.02254000 1.08262000 C - 0.00894000 - 0.01624000 - 0.43421000 H - 0.49334000 0.93505000 1.44716000 H 1.05522000 0.04512000 1.44808000 H - 0.64695000 - 1.12346000 2.54219000 H 0.50112000 - 0.91640000 - 0.80440000 H 0.49999000 0.86726000 - 0.84481000 H - 1.04310000 - 0.02739000 - 0.80544000 O - 0.66442000 - 1.15471000 1.56909000
-
Use the File → New menu command in AMSinput
Copy the xyz coordinates of ethanol on this page (in your browser)
Click in the AMSinput window to activate it
Paste the xyz coordinates (ctrl/cmd-V or Edit → Paste)

You should again get the ethanol molecule, just as you have saved it.
Import SMILES string¶
AMSinput can also interpret SMILES strings (via OpenBabel). As a demonstration, lets try again with Ethanol:
-
Use the File → New menu command in AMSinput
Click 'No' when asked if you want to save your changes
Open a web browser
Search for ethanol on wikipedia.org
At the right side of the page, click the 'Show' link to show the SMILES
Copy the SMILES string ( CCO )
Click in the AMSinput window to activate it
Paste the SMILES string (ctrl/cmd-V or Edit → Paste)
Click in empty space in the drawing area to clear the selection

Again we have an ethanol molecule. SMILES strings do not contain the 3D structure, it was generated by OpenBabel and is NOT an ADF optimized structure. So normally the next step would be to pre-optimize with UFF (via the cog wheel), and to optimize the geometry with ADF.
Your own structures library¶
You can make your own structure library very easily.
By default, user defined structures will be stored in the .scm_gui/Structures directory.
Defining your structures¶
To be able to actually use the structures as described earlier, it is necessary to define one of the atoms as having xyz-coordinates (0,0,0). This will then be the atom that will actually appear at the spot of the atom that is replaced by the structure. If you use the Save As Structure command this will be done for you.
-
Select the File → New command
Click No (do not save changes)
Build methane
Replace three of the hydrogens by chloride atoms and pre-optimize
Delete the remaining hydrogen
Select the central carbon atom
Use the Structures tool → Save As Structure … command
Enter a name like trichloromethyl
Note that the selected atom (currently the C atom) will be used as anchor
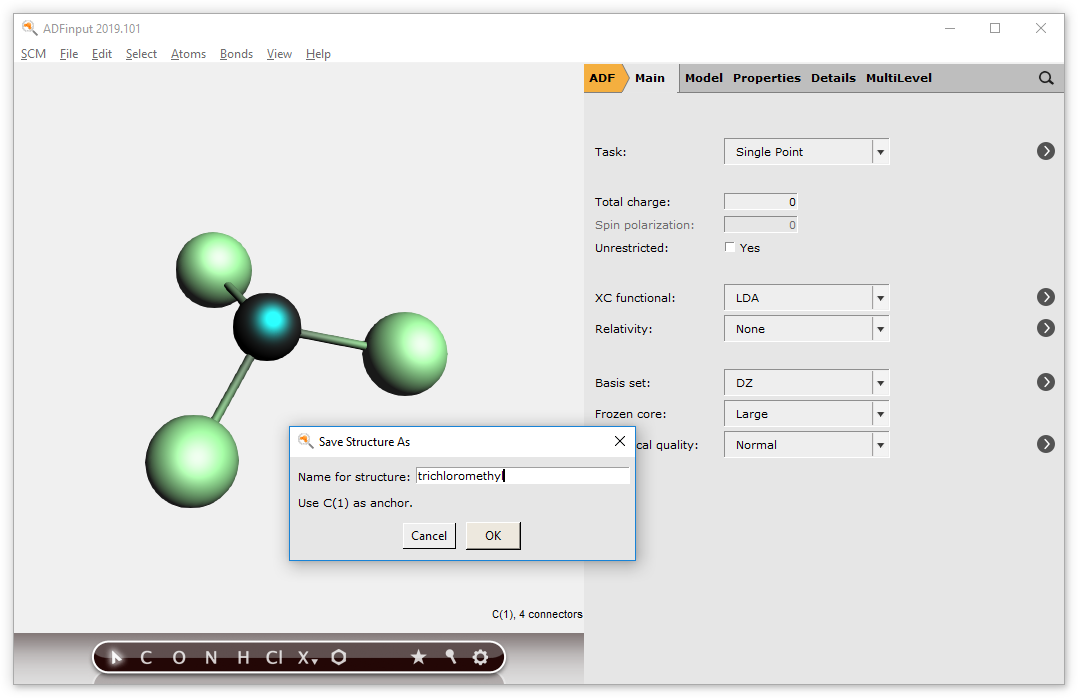
The new structure will appear in the structures menu and can be directly used.
Using dummy atoms¶
Dummy ("Xx") atoms are treated differently when used in structures. A dummy atom will not replace an existing atom when it is defined as the 'replacing atom'. Instead, the double-clicked atom will remain and will accept the bonds that the dummy atom had in the structure.
-
Build a methane molecule
Replace the carbon atom by a nitrogen atom
Select one of the hydrogens and replace it by a dummy atom (the Xx atom type, in the periodic system)
Select the dummy atom
Save the structure using the Structures tool → Save As Structure … command

Now you can select the structure from the structures menu and directly use it.
-
Select your new structure from the structures menu
Double-click on one of the hydrogens
Notice that the hydrogen atom is not removed and that the NH3 group is attached to it. Similar behavior has been demonstrated with the bidentate ligands, where the dummy atoms are also used.
If you want to clean up your structures, you can use the Structures tool → Manage Structures… command. If you use it, AMSjobs will open and show the contents of your Structures directory. As the structures are just (simplified) .ams files, you can open them using AMSinput. And using AMSjobs you can rename them or delete them.
A sphere of Cu atoms, cut out of the crystal¶
We start making a Cu crystal, using a super cell so we have many real Cu atoms.
To build the crystal, we need to use the periodic tools. These will work only for programs supporting periodicity.
-
Start AMSinput (or use File → New in the currently open AMSinput window)
Search (
 ) for 'copper'
) for 'copper'Click on 'Cu' in the Crystals section of the search results
Edit → Crystal → Generate Super Cell…
Enter '5' to change left top element to 5 (the other diagonal elements should automatically adjust)
Click OK in the pop-up-window
Uncheck View → Periodic → Repeat Unit Cells, so only one unit cell will be shown
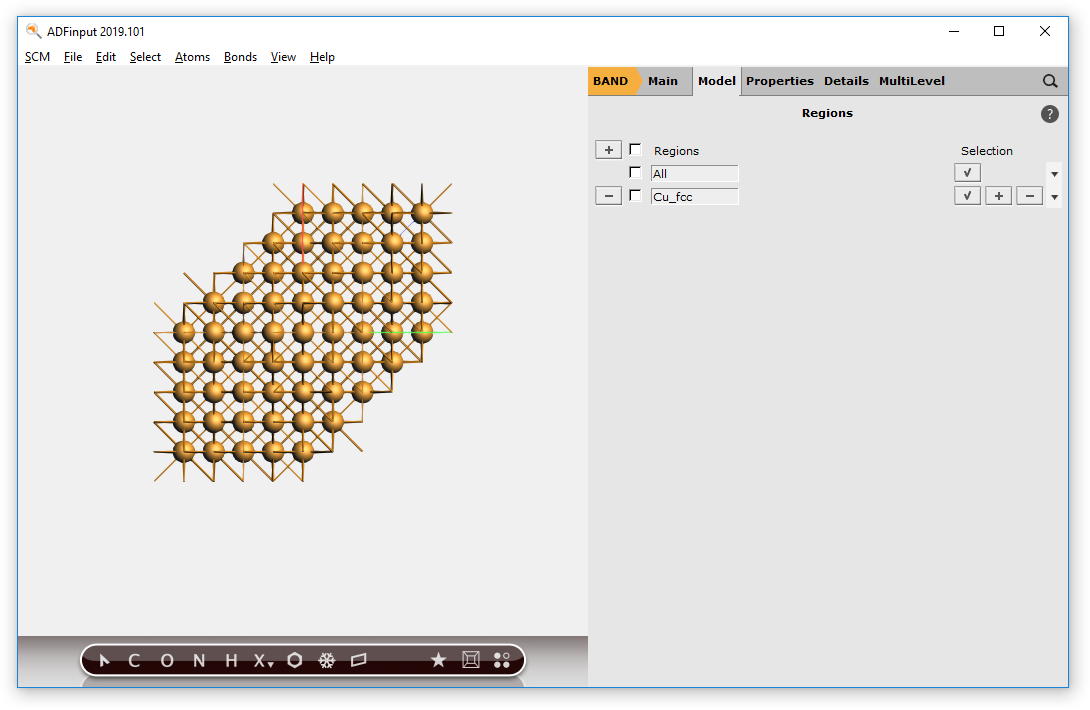
Now we have a block of Cu, with explicit Cu atoms (that is using a super cell). Next we will center this block, and select a sphere of atoms around the origin.
-
Make sure the origin is in the center of the block: Edit → Set Origin
Select → Select Atom Close To Origin
Select → Select Within Radius
Click OK

-
Select → Invert Selection
Press the Backspace key to delete the selected atoms
If it does not respond: click once in the drawing area to focus on it, and press the Backspace key again
Switch to ADF: panel bar BAND → ADF (BAND might be a different like DFTB, ReaxFF or UFF)
Rotate a little
As you can see, you have a (very small) sphere consisting of Cu atoms in the molecular ADF program:
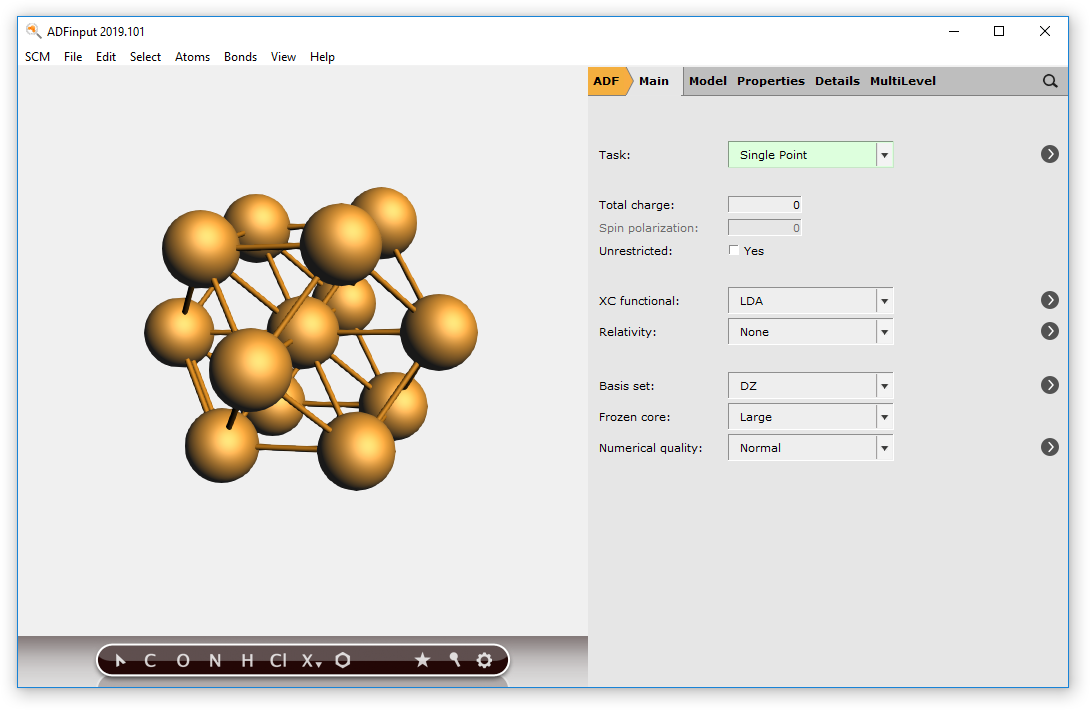
Obviously, by making a bigger super-cell and selecting atoms within a larger radius you can make bigger spheres.
A carbon nanotube¶
A small piece of nanotube is included in the molecule database, so you can just search for it and use it. However, typically one wants some specific nanotube structure. And make it infinite (periodic in one dimension). This can conveniently be done by importing the structure as found on the web:
-
Request CIF format as output, leave other options at the default values
Click generate
In the browser windows we get the nanotube structure in CIF format, something like the following:
data_nanotube _audit_creation_method '(3,3) Nanotube -- TubeGen 3.3, J T Frey, University of Delaware' _cell_length_a 7.4762 _cell_length_b 7.4762 _cell_length_c 2.4643 _cell_angle_alpha 90.00 _cell_angle_beta 90.00 _cell_angle_gamma 120.00 _symmetry_space_group_name_H - M 'P 1' _symmetry_Int_Tables_number 1 loop_ _atom_site_label _atom_site_fract_x _atom_site_fract_y _atom_site_fract_z C 0.7762 0.5000 0.0000 C 0.8138 0.7061 0.0000 C 0.7762 0.7762 0.5000 C 0.6077 0.8138 0.5000 C 0.5000 0.7762 0.0000 C 0.2939 0.6077 0.0000 C 0.2238 0.5000 0.5000 C 0.1862 0.2939 0.5000 C 0.2238 0.2238 0.0000 C 0.3923 0.1862 0.0000 C 0.5000 0.2238 0.5000 C 0.7061 0.3923 0.5000
Next we want to get this structure into AMSinput:
-
Select the CIF information in your browser
Copy
Start AMSinput
Edit → Paste
View → Axes
Edit → Set Origin
View → Periodic → Repeat Unit Cells
Rotate to get a good view

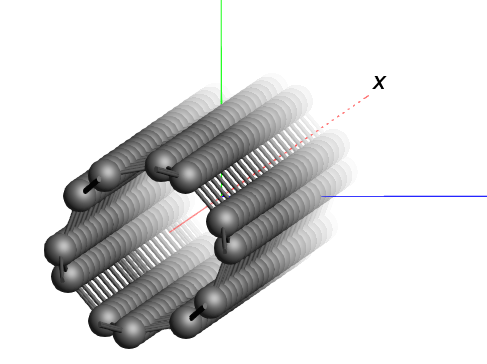
We see a piece of nanotube, repeated in all directions. Actually nine nanotubes are visible.
Notice that the nanotubes are oriented along the Z-axes.
The GUI can handle one-dimensional systems. However, in case of a one dimensional system (a chain) the lattice vector is always along the X direction. So to change our nanotube in a nice periodic one-dimensional structure we need to rotate it (including the lattice vectors) such that the tube is along the X-axes.
-
Use the Edit → Rotate 90 menu command to make the nanotube lie along the X-axes (hint: rotate around the Y-axes)
Switch to a one-dimensional system (chain)

The Rotate 90 command did not only rotate the coordinates of the atoms, but also the lattice vectors. Now we have a small piece of nanotube (remember it already is repeated 3 times for visualization purposes). This might be sufficient for your purposes, but it is easy to make a bigger piece:
-
Edit → Crystal → Generate Super Cell
Enter 10 in the topleft cell
Click OK to repeat the unit cell 10 times

Switch to some non-periodic code (like ADF) if you wish to treat this piece of nanotube without infinite symmetry.
If you have a large system you can sometimes get a better view by introducing Fog. This works best with a white background.
-
View → Background → White
View → Fog
Click the Done button (or play with the sliders first if you want to change the fog parameters)
Draw The Structure Of Ethanol Molecule
Source: https://www.scm.com/doc/Tutorials/BuildingStructures/BuildingMolecules.html
Posted by: hubbardwhationam.blogspot.com

0 Response to "Draw The Structure Of Ethanol Molecule"
Post a Comment